知识图谱嵌入模型之TransE算法
TransE
·
知识图谱嵌入
知识图谱是一个三元组组成的集合,将头尾实体通过关系连接成一个图,而知识图谱存在一个问题,就是离散的图结构是不能够进行语义计算的,为帮助计算机对知识进行计算,解决数据稀疏性,可以将知识图谱中的实体、关系映射到低维连续的向量空间中,这类方法称为知识图谱嵌入。
TransE
TransE提出了一种将实体与关系嵌入到低维向量空间中的简单模型
原理:
TransE基于实体和关系的分布式向量表示,将每个三元组实例(head,relation,tail)中的关系relation看做从实体head到实体tail的翻译,即向量相加,通过不断调整h、r和t的向量,使(h + r) 尽可能与 t 相等,即,头实体向量 + 关系向量 = 尾实体向量 (h + r = t)。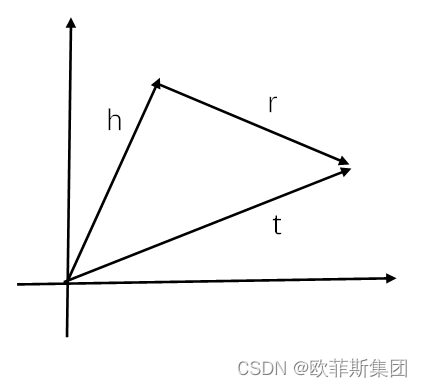
TransE代码
import codecs
import numpy as np
import copy
import time
import random
def dataloader(file1, file2, file3):
"""
加载数据
file1:训练数据集
file2:实体集合
file3:关系集合
返回数据:三个列表,实体,关系,三元组
"""
entity = []
relation = []
entities2id = {}
relations2id = {}
with open(file2, 'r') as f1, open(file3, 'r') as f2:
lines1 = f1.readlines()
lines2 = f2.readlines()
for line in lines1:
line = line.strip().split('\t')
if len(line) != 2:
continue
entities2id[line[0]] = line[1]
entity.append(line[1])
for line in lines2:
line = line.strip().split('\t')
if len(line) != 2:
continue
relations2id[line[0]] = line[1]
relation.append(line[1])
triple_list = []
with codecs.open(file1, 'r') as f:
content = f.readlines()
for line in content:
triple = line.strip().split("\t")
if len(triple) != 3:
continue
h_ = entities2id[triple[0]]
r_ = relations2id[triple[1]]
t_ = entities2id[triple[2]]
triple_list.append([h_, r_, t_])
print("Complete load. entity : %d , relation : %d , triple : %d" % (
len(entity), len(relation), len(triple_list)))
return entity, relation, triple_list
class TransE:
def __init__(self, entity, relation, triple_list, embedding_dim=50, lr=0.01, margin=1.0, norm=1):
"""参数初始化"""
self.entities = entity
self.relations = relation
self.triples = triple_list
self.dimension = embedding_dim
self.learning_rate = lr
self.margin = margin
self.norm = norm
self.loss = 0.0
def data_initialise(self):
"""将实体id列表、关系id列表转变为entityVectorList和relationVectorList两个字典。"""
entityVectorList = {} # {实体id:实体向量}
relationVectorList = {} # {关系id:关系向量}
for entity in self.entities:
entity_vector = np.random.uniform(-6.0 / np.sqrt(self.dimension), 6.0 / np.sqrt(self.dimension),self.dimension)
entityVectorList[entity] = entity_vector
for relation in self.relations:
relation_vector = np.random.uniform(-6.0 / np.sqrt(self.dimension), 6.0 / np.sqrt(self.dimension),self.dimension)
relation_vector = self.normalization(relation_vector)
relationVectorList[relation] = relation_vector
self.entities = entityVectorList
self.relations = relationVectorList
def normalization(self, vector):
return vector / np.linalg.norm(vector)
def training_run(self, epochs=1, nbatches=100, out_file_title = ''):
batch_size = int(len(self.triples) / nbatches)
print("batch size: ", batch_size)
for epoch in range(epochs):
start = time.time()
self.loss = 0.0
# Normalise the embedding of the entities to 1
for entity in self.entities.keys():
self.entities[entity] = self.normalization(self.entities[entity]);
for batch in range(nbatches):
batch_samples = random.sample(self.triples, batch_size)
Tbatch = []
for sample in batch_samples:
corrupted_sample = copy.deepcopy(sample)
pr = np.random.random(1)[0]
if pr > 0.5:
# change the head entity
corrupted_sample[0] = random.sample(self.entities.keys(), 1)[0]
while corrupted_sample[0] == sample[0]:
corrupted_sample[0] = random.sample(self.entities.keys(), 1)[0]
else:
# change the tail entity
corrupted_sample[2] = random.sample(self.entities.keys(), 1)[0]
while corrupted_sample[2] == sample[2]:
corrupted_sample[2] = random.sample(self.entities.keys(), 1)[0]
if (sample, corrupted_sample) not in Tbatch:
Tbatch.append((sample, corrupted_sample))
self.update_triple_embedding(Tbatch)
end = time.time()
print("epoch: ", epoch, "cost time: %s" % (round((end - start), 3)))
print("running loss: ", self.loss)
with codecs.open(out_file_title +"TransE_entity_" + str(self.dimension) + "dim_batch" + str(batch_size), "w") as f1:
for e in self.entities.keys():
f1.write(e + "\t")
f1.write(str(list(self.entities[e])))
f1.write("\n")
with codecs.open(out_file_title +"TransE_relation_" + str(self.dimension) + "dim_batch" + str(batch_size), "w") as f2:
for r in self.relations.keys():
f2.write(r + "\t")
f2.write(str(list(self.relations[r])))
f2.write("\n")
def update_triple_embedding(self, Tbatch):
copy_entity = copy.deepcopy(self.entities)
copy_relation = copy.deepcopy(self.relations)
for correct_sample, corrupted_sample in Tbatch:
correct_copy_head = copy_entity[correct_sample[0]]
correct_copy_tail = copy_entity[correct_sample[2]]
relation_copy = copy_relation[correct_sample[1]]
corrupted_copy_head = copy_entity[corrupted_sample[0]]
corrupted_copy_tail = copy_entity[corrupted_sample[2]]
correct_head = self.entities[correct_sample[0]]
correct_tail = self.entities[correct_sample[2]]
relation = self.relations[correct_sample[1]]
corrupted_head = self.entities[corrupted_sample[0]]
corrupted_tail = self.entities[corrupted_sample[2]]
# calculate the distance of the triples
if self.norm == 1:
correct_distance = norm_l1(correct_head, relation, correct_tail)
corrupted_distance = norm_l1(corrupted_head, relation, corrupted_tail)
else:
correct_distance = norm_l2(correct_head, relation, correct_tail)
corrupted_distance = norm_l2(corrupted_head, relation, corrupted_tail)
loss = self.margin + correct_distance - corrupted_distance
if loss > 0:
self.loss += loss
correct_gradient = 2 * (correct_head + relation - correct_tail)
corrupted_gradient = 2 * (corrupted_head + relation - corrupted_tail)
if self.norm == 1:
for i in range(len(correct_gradient)):
if correct_gradient[i] > 0:
correct_gradient[i] = 1
else:
correct_gradient[i] = -1
if corrupted_gradient[i] > 0:
corrupted_gradient[i] = 1
else:
corrupted_gradient[i] = -1
correct_copy_head -= self.learning_rate * correct_gradient
relation_copy -= self.learning_rate * correct_gradient
correct_copy_tail -= -1 * self.learning_rate * correct_gradient
relation_copy -= -1 * self.learning_rate * corrupted_gradient
if correct_sample[0] == corrupted_sample[0]:
# if corrupted_triples replaces the tail entity, the head entity's embedding need to be updated twice
correct_copy_head -= -1 * self.learning_rate * corrupted_gradient
corrupted_copy_tail -= self.learning_rate * corrupted_gradient
elif correct_sample[2] == corrupted_sample[2]:
# if corrupted_triples replaces the head entity, the tail entity's embedding need to be updated twice
corrupted_copy_head -= -1 * self.learning_rate * corrupted_gradient
correct_copy_tail -= self.learning_rate * corrupted_gradient
# normalising these new embedding vector, instead of normalising all the embedding together
copy_entity[correct_sample[0]] = self.normalization(correct_copy_head)
copy_entity[correct_sample[2]] = self.normalization(correct_copy_tail)
if correct_sample[0] == corrupted_sample[0]:
# if corrupted_triples replace the tail entity, update the tail entity's embedding
copy_entity[corrupted_sample[2]] = self.normalization(corrupted_copy_tail)
elif correct_sample[2] == corrupted_sample[2]:
# if corrupted_triples replace the head entity, update the head entity's embedding
copy_entity[corrupted_sample[0]] = self.normalization(corrupted_copy_head)
# the paper mention that the relation's embedding don't need to be normalised
copy_relation[correct_sample[1]] = relation_copy
# copy_relation[correct_sample[1]] = self.normalization(relation_copy)
self.entities = copy_entity
self.relations = copy_relation
if __name__ == '__main__':
file1 = "./train.txt"
file2 = "./entity2id.txt"
file3 = "./relation2id.txt"
entity_set, relation_set, triple_list = dataloader(file1, file2, file3)
transE = TransE(entity_set, relation_set, triple_list, embedding_dim=50, lr=0.01, margin=1.0, norm=2)
transE.data_initialise()
transE.training_run(out_file_title="test")

魔乐社区(Modelers.cn) 是一个中立、公益的人工智能社区,提供人工智能工具、模型、数据的托管、展示与应用协同服务,为人工智能开发及爱好者搭建开放的学习交流平台。社区通过理事会方式运作,由全产业链共同建设、共同运营、共同享有,推动国产AI生态繁荣发展。
更多推荐
 已为社区贡献3条内容
已为社区贡献3条内容







所有评论(0)